My research focuses on the intersection of computational pathology and artificial intelligence for improving cancer diagnosis and patient outcomes.
Specifically, I am interested in developing machine learning methods for Digital Pathology with a specific focus on Self-supervised, Semi-supervised and Weakly-supervised learning.
|
Self-Supervised Driven Consistency Training for Annotation Efficient Histopathology Image Analysis
Chetan L Srinidhi, Seung Wook Kim, Fu-Der Chen, Anne L Martel
Medical Image Analysis (MedIA), 2022
(Top Journal – IF: 8.545)
[Paper]
[Code]
[Bibtex]
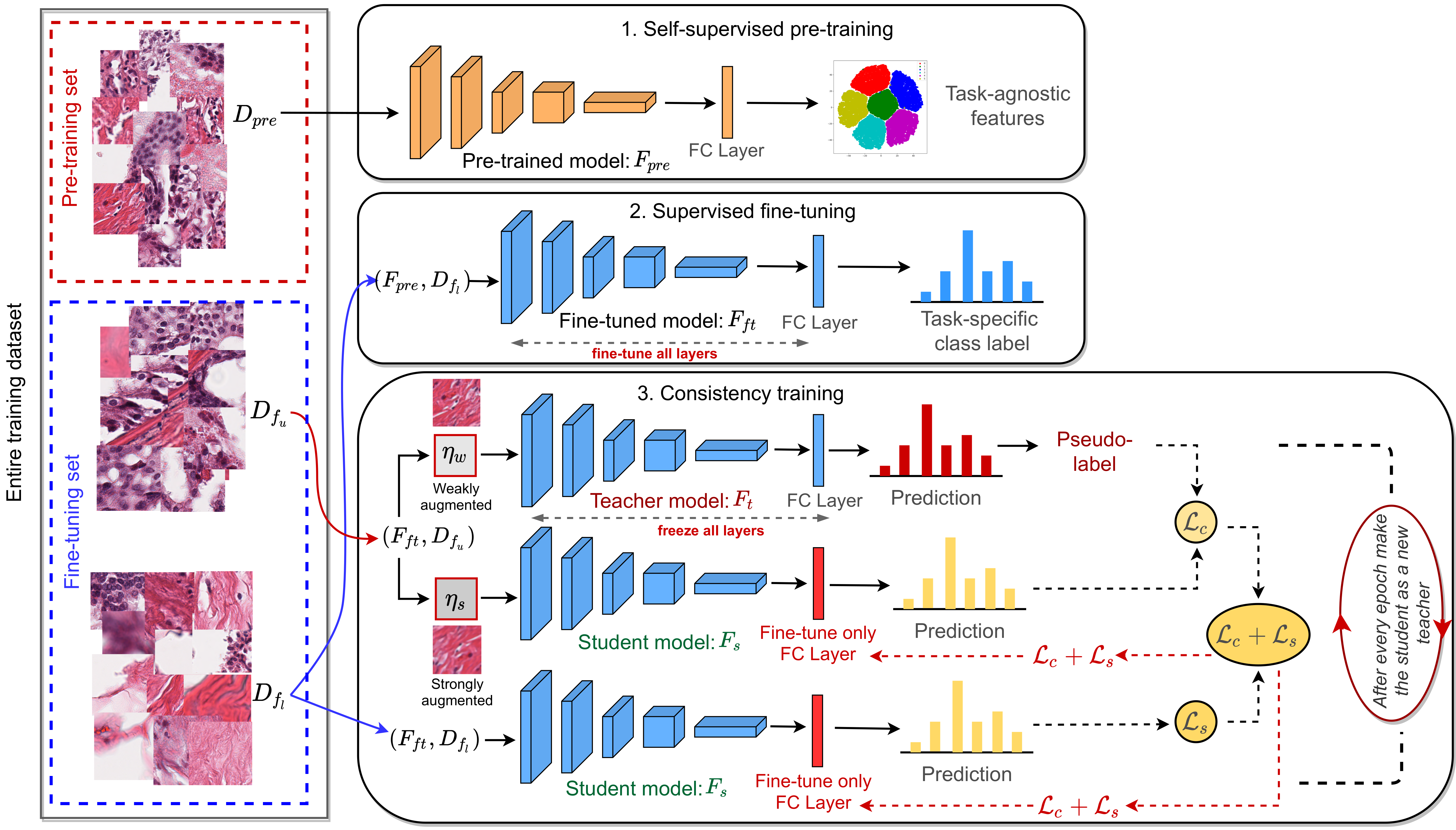
Training a neural network with a large labeled dataset is still a dominant paradigm in computational histopathology. However, obtaining such exhaustive manual annotations is often expensive, laborious, and prone to inter and intra-observer variability. While recent self-supervised and semi-supervised methods can alleviate this need by learning unsupervised feature representations, they still struggle to generalize well to downstream tasks when the number of labeled instances is small.
In this work, we overcome this challenge by leveraging both task-agnostic and task-specific unlabeled data based on two novel strategies: i) a self-supervised pretext task that harnesses the underlying multi-resolution contextual cues in histology whole-slide images to learn a powerful supervisory signal for unsupervised representation learning; ii) a new teacher-student semi-supervised consistency paradigm that learns to effectively transfer the pretrained representations to downstream tasks based on prediction consistency with the task-specific unlabeled data.
We carry out extensive validation experiments on three histopathology benchmark datasets across two classification and one regression based tasks, i.e., tumor metastasis detection, tissue type classification, and tumor cellularity quantification. Under limited label data, the proposed method yields tangible improvements, which is close or even outperforming other state-of-the-art self-supervised and supervised baselines. Furthermore, we empirically show that the idea of bootstrapping the self-supervised pretrained features is an effective way to improve the task-specific semi-supervised learning on standard benchmarks.
|
|
|
|
|
|
|
|
|
|
|
Consistency driven Sequential Transformers Attention Model for Partially Observable Scenes
Samrudhdhi B Rangrej, Chetan L Srinidhi, James J Clark
IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2022, (Accepted)
[Paper]
[Code]
[Bibtex]
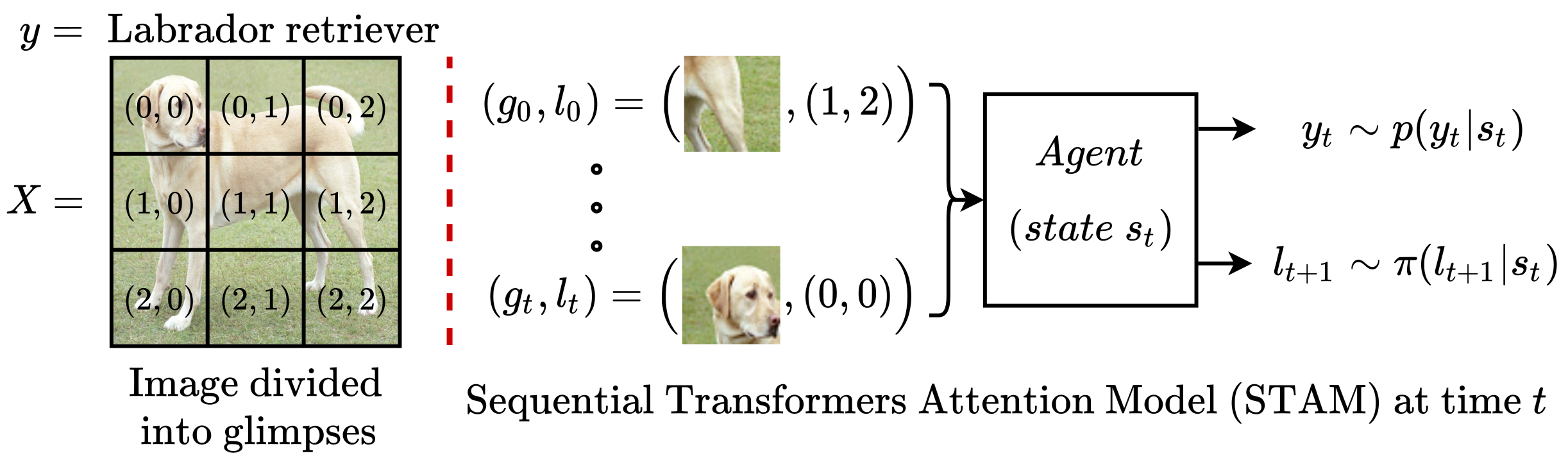
Most hard attention models initially observe a complete scene to locate and sense informative glimpses, and predict class-label of a scene based on glimpses.
However, in many applications (e.g., aerial imaging), observing an entire scene is not always feasible due to the limited time and resources available for acquisition. In this paper, we develop a Sequential Transformers Attention Model (STAM) that only partially observes a complete image and predicts informative glimpse locations solely based on past glimpses.
We design our agent using DeiT-distilled and train it with a one-step actor-critic algorithm. Furthermore, to improve classification performance, we introduce a novel training objective, which enforces consistency between the class distribution predicted by a teacher model from a complete image and the class distribution predicted by our agent using glimpses. When the agent senses only 4% of the total image area, the inclusion of the proposed consistency loss in our training objective yields 3% and 8% higher accuracy on ImageNet and fMoW datasets, respectively.
Moreover, our agent outperforms previous state-of-the-art by observing nearly 27% and 42% fewer pixels in glimpses on ImageNet and fMoW.
|
|
|
|
|
|
|
|
|
|
|
Improving Self-Supervised Learning with Hardness-aware Dynamic Curriculum Learning: An Application to Digital Pathology
Chetan L Srinidhi, Anne L Martel
International Conference on Computer Vision (ICCV), 2021, CDpath Workshop (Oral)
[Paper]
[Code]
[Bibtex]
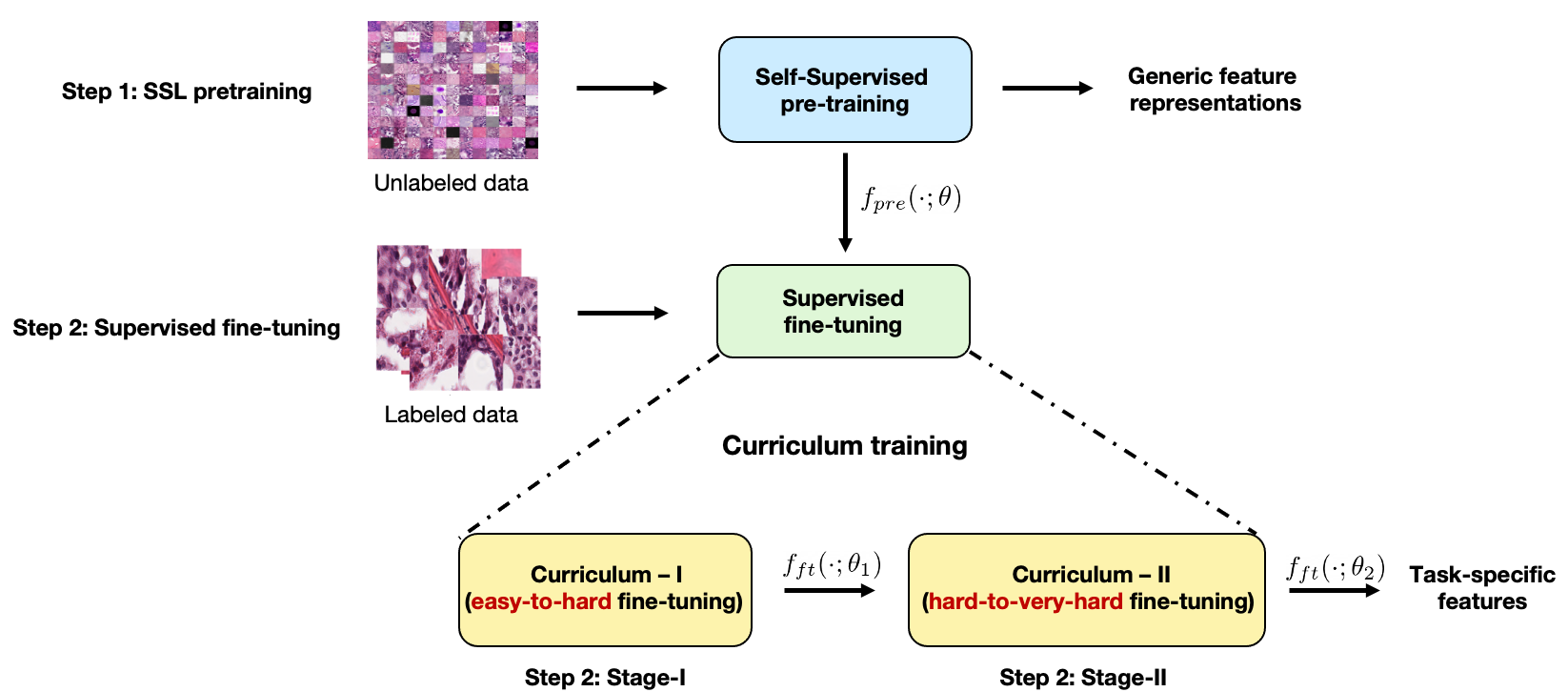
Self-supervised learning (SSL) has recently shown tremendous potential to learn generic visual representations useful for many image analysis tasks. Despite their notable success, the existing SSL methods fail to generalize to downstream tasks when the number of labeled training instances is small or if the domain shift between the transfer domains is significant.
In this paper, we attempt to improve self-supervised pretrained representations through the lens of curriculum learning by proposing a hardness-aware dynamic curriculum learning (HaDCL) approach. To improve the robustness and generalizability of SSL, we dynamically leverage progressive harder examples via easy-to-hard and hard-to-very-hard samples during mini-batch downstream fine-tuning. We discover that by progressive stage-wise curriculum learning, the pretrained representations are significantly enhanced and adaptable to both in-domain and out-of-domain distribution data.
We performed extensive validation on three histology benchmark datasets on both patch-wise and slide-level classification problems. Our curriculum based fine-tuning yields a significant improvement over standard fine- tuning, with a minimum improvement in area-under-the-curve (AUC) score of 1.7% and 2.2% on in-domain and out-of-domain distribution data, respectively. Further, we empirically show that our approach is more generic and adaptable to any SSL methods and does not impose any additional overhead complexity. Besides, we also outline the role of patch-based versus slide-based curriculum learning in histopathology to provide practical insights into the success of curriculum based fine-tuning of SSL methods.
|
|
|
|
|
|
|
|
|
|
|
Deep Neural Network Models for Computational Histopathology: A Survey
Chetan L Srinidhi, Ozan Ciga, Anne L Martel
Medical Image Analysis (MedIA), 2020
(Top Journal – IF: 8.545)
Most downloaded article in MedIA
[Paper]
[Bibtex]
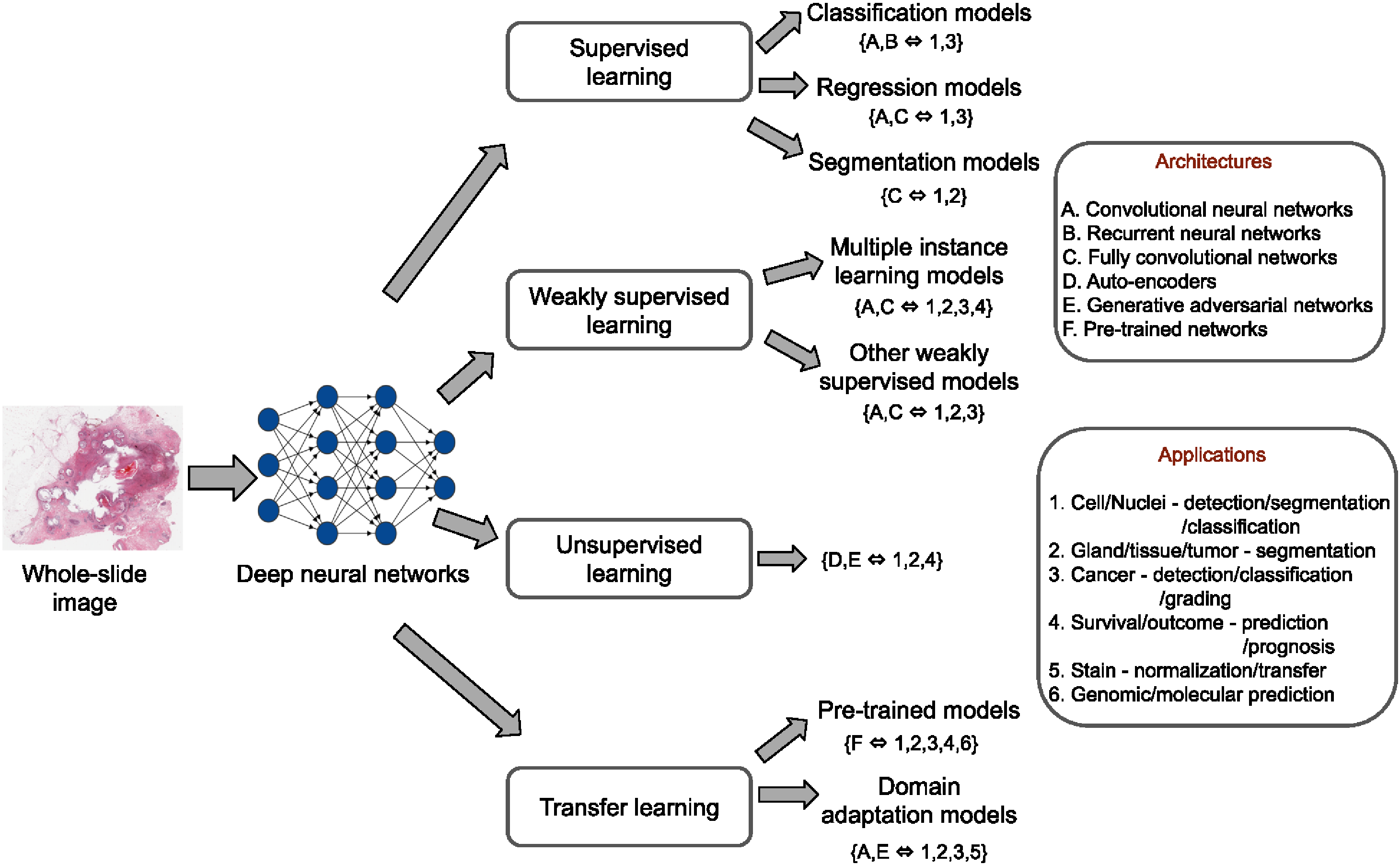
In this paper, we present a comprehensive review of state-of-the-art deep learning approaches that have been used in the context of histopathological image analysis. From the survey of over 130 papers, we review the field’s progress based on the methodological
aspect of different machine learning strategies such as supervised, weakly supervised, unsupervised, transfer learning
and various other sub-variants of these methods. We also provide an overview of deep learning based survival models that are applicable for disease-specific prognosis tasks. Finally, we summarize several existing open datasets and
highlight critical challenges and limitations with current deep learning approaches, along with possible avenues for future research.
|
|
|
|
|
|
|
|
|
|
|
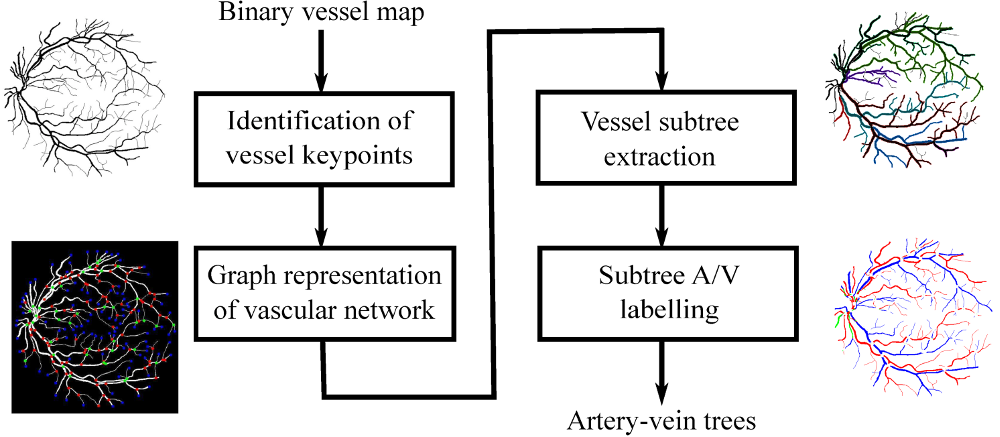
Automated Method for Retinal Artery/Vein Separation via Graph Search Metaheuristic Approach
Chetan L Srinidhi, P Aparna, and Jeny Rajan
IEEE Transactions on Image Processing (TIP), 2019
(Top Journal – IF: 10.856)
[Paper]
[Bibtex]
In this paper, we present a novel graph search metaheuristic approach for automatic separation of arteries/veins (A/V) from color fundus images.
Our method exploits local information to disentangle the complex vascular tree into multiple subtrees, and global information to label these vessel subtrees into arteries and veins.
Given a binary vessel map, a graph representation of the vascular network is constructed representing the topological and spatial connectivity of the vascular structures.
Based on the anatomical uniqueness at vessel crossing and branching points, the vascular tree is split into multiple subtrees containing arteries and veins.
Finally, the identified vessel subtrees are labeled with A/V based on a set of handcrafted features trained with random forest classifier.
The proposed method has been tested on four different publicly available retinal datasets with an average accuracy of 94.7%, 93.2%, 96.8% and 90.2% across AV-DRIVE, CT-DRIVE. INSPIRE-AVR and WIDE datasets, respectively.
These results demonstrate the superiority of our proposed approach in outperforming state-of-the- art methods for A/V separation.
|
|
|
|
|
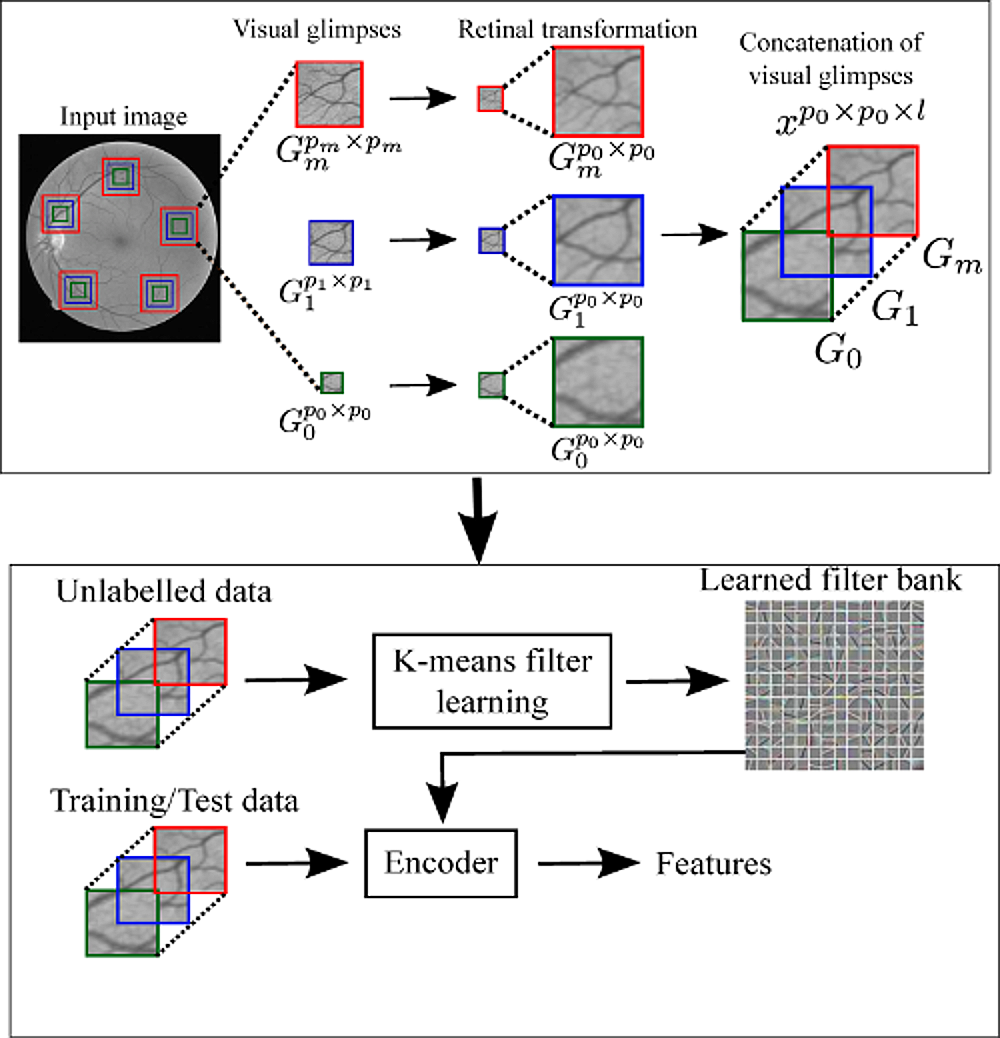
A Visual Attention Guided Unsupervised Feature Learning for Robust Vessel Delineation in Retinal Images
Chetan L Srinidhi, P Aparna, and Jeny Rajan
Biomedical Signal Processing and Control, 2018
(Journal – IF: 3.88)
[Paper]
[Bibtex]
We propose a novel visual attention guided unsupervised feature learning (VA-UFL) approach to automatically learn the most discriminative features for segmenting vessels in retinal images.
Our VA-UFL approach captures both the knowledge of visual attention mechanism and multi-scale contextual information to selectively visualize the most relevant part of the structure in a given local patch.
This allows us to encode a rich hierarchical information into unsupervised filtering learning to generate a set of most discriminative features that aid in the accurate segmentation of vessels, even in the presence of cluttered background.
Our proposed method is validated on the five publicly available retinal datasets: DRIVE, STARE, CHASE_DB1, IOSTAR and RC-SLO.
The experimental results show that the proposed approach significantly outperformed the state-of-the-art methods in terms of sensitivity, accuracy and area under the receiver operating characteristic curve across all five datasets.
Specifically, the method achieved an average sensitivity greater than 0.82, which is 7% higher compared to all existing approaches validated on DRIVE, CHASE_DB1, IOSTAR and RC-SLO datasets, and outperformed even second-human observer.
The method is shown to be robust to segmentation of thin vessels, strong central vessel reflex, complex crossover structures and fares well on abnormal cases.
|
|
|
|
|
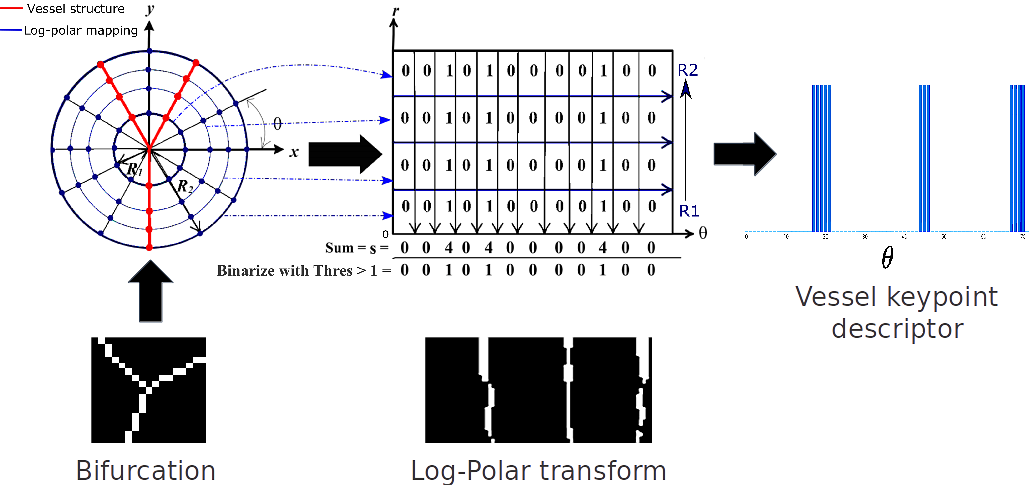
A Vessel Keypoint Detector for Junction classification
Chetan L Srinidhi, Priyadarshi Rath, Jayanthi Sivaswamy
IEEE International Symposium on Biomedical Imaging (ISBI), 2017
(Oral Presentation, Acceptance Rate: ~19%)
[Paper]
[Bibtex]
In this paper, we propose a novel Vessel Keypoint Detector (VKD) which is derived from the projection of log-polar transformed binary patches around vessel points.
VKD is used to design a two stage solution for junction detection and classification. In the first stage, the keypoints detected using VKD are refined using curvature orientation information to extract candidate junctions.
True junctions from these candidates are identified in a supervised manner using a Random Forest classifier.
In the next stage, a novel combination of local orientation and shape based features is extracted from the junction points and classified using a second Random Forest classifier.
Evaluation results on five datasets show that the designed system is robust to changes in resolution and other variations across datasets, with average values of accuracy/sensitivity/specificity for junction detection being 0.78/0.79/0.75 and for junction classification being 0.87/0.85/0.88.
Our system outperforms the state of the art method by at least 11%, on the DRIVE and IOSTAR datasets. These results demonstrate the effectiveness of VKD for vessel analysis.
|
|
|
|
|
|
|
|
|
|
|
|
Recent Advancements in Retinal Vessel Segmentation
Chetan L Srinidhi, P Aparna, and Jeny Rajan
Journal of Medical Systems, 2017
(Journal – IF: 4.460)
[Paper] [Bibtex]
In this paper, we carry out a systematic review of the most recent advancements in retinal vessel segmentation methods published in last five years. The objectives of this study are as follows:
first, we discuss the most crucial preprocessing steps that are involved in accurate segmentation of vessels. Second, we review most recent state-of-the-art retinal vessel segmentation techniques which are classified into different categories based on their main principle.
Third, we quantitatively analyse these methods in terms of its sensitivity, specificity, accuracy, area under the curve and discuss newly introduced performance metrics in current literature. Fourth, we discuss the advantages and limitations of the existing segmentation techniques.
Finally, we provide an insight into active problems and possible future directions towards building successful computer-aided diagnostic system.
|







